
Preventing PCR Contamination One Reaction at a Time
Polymerase chain reaction or PCR was first described in its modern form in 1985. Today, PCR is one of the most widely used laboratory techniques for DNA amplification and has become a valuable tool across multiple disciplines from infectious disease and molecular biology to paleontology. Many credit PCR as one of the major advancements required for the birth of biotechnology. Modifications to the original workflow and advances in technology has not only improved PCR performance, but led to the development of techniques for DNA quantitation (qPCR and digital droplet PCR) and RNA amplification (RT-PCR) and quantitation (qRT-PCR) as well.
The Steps of PCR
The utility of PCR, in any form, stems from its specificity, efficiency, and sensitivity. Specificity in PCR reactions is dictated by the sequence of primers or short single-stranded pieces of DNA that are complementary to the beginning and/or end portions of the target DNA sequence to be amplified or copied. Primers are utilized in pairs such that one is complementary to and binds to the beginning of the target DNA sequence and the second binds to the end. To prepare a reaction for PCR, primer pairs are mixed with the DNA sample to be amplified, free oligonucleotides, and DNA polymerase enzyme in a buffered salt solution. Once mixed, amplification of the target DNA sequence can begin using a 3-step temperature-dependent cycle (Figure 1). The first PCR cycle is started by heating the reaction to a high temperature, which denatures or separates the two strands of sample DNA. The reaction is then cooled to allow the primers to bind or anneal to the complimentary sequences within the sample DNA. The annealed primers create a small double-stranded portion of DNA that acts as a scaffold for DNA polymerase binding. Once bound, DNA polymerase adds oligonucleotides to the end of each annealed primer using the single-stranded target DNA as a template. This effectively creates a total of two copies of the target DNA at the end of the first PCR cycle. This cycle is typically repeated 30-40 times to create millions of copies of the target DNA within a few hours.
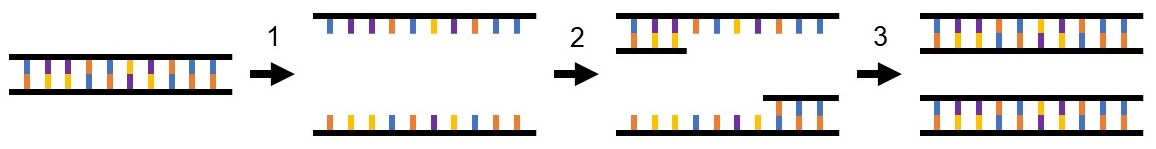 Figure 1. PCR amplifies DNA in a three-step process. 1) Denaturation: double-stranded DNA from the sample is denatured into two single strands, 2) Annealing: the complementary primer pair binds to the target DNA sequence within the sample DNA, 3) Extension: DNA polymerase adds free oligonucleotides to the end of each primer to create two double-stranded copies of the target DNA sequence.
Figure 1. PCR amplifies DNA in a three-step process. 1) Denaturation: double-stranded DNA from the sample is denatured into two single strands, 2) Annealing: the complementary primer pair binds to the target DNA sequence within the sample DNA, 3) Extension: DNA polymerase adds free oligonucleotides to the end of each primer to create two double-stranded copies of the target DNA sequence.
Sensitivity: A Double-edged Sword
The ability to create millions of copies of a single DNA/RNA molecule makes PCR a powerful and sensitive tool. However, it also makes it susceptible to contaminating environmental DNA, which can be amplified during the PCR workflow in the same manner as the target molecule. The need for annealed primers for DNA polymerase binding lends some protection from amplification of contaminating DNA, but primer binding does not require perfect complementarity to a DNA sequence for annealing to occur. Mismatches between the primer and target sequence will lead to less efficient primer binding and subsequently less efficient amplification, but even inefficient background amplification can lead to issues with downstream sample analyses, especially when quantitation is required. Environmental, operator, and/or reagent contamination often leads to false positives in qPCR or qRT-PCR experiments, making interpretation of the results nearly impossible. An added challenge associated with RNA amplification and quantitation is RNase contamination. RNase, an enzyme that degrades RNA, is prevalent in on the human skin and lab environment; contaminating lab benches, solutions, pipets etc. RNA samples contaminated with RNase can be completely degraded within minutes rendering the sample unusable. Together these challenges highlight the need to include proper experimental controls and prevent sample and PCR contamination to obtain accurate results.
Preventing PCR Contamination
When preparing PCR samples, operators should always don the appropriate personal protective equipment to protect themselves, as well as the sample. Sample preparation, nucleotide isolation, and PCR reaction mixing should always be carried out in a clean area using reagents suitable for molecular biology applications (e.g. RNase and DNase free). To prevent contamination from the lab environment, reactions can be set up in a laminar flow hood with a vertical laminar flow. The hood draws in air from the top of the hood and through a high efficiency particulate air (HEPA) filter. The filtered air then flows down to the workspace and finally out through the front sash near the operator. Alternatively, a dead air box which provides an area of circulation-free air can be used to prevent contamination of PCR samples. It is important to note that dead air boxes and laminar flow hoods provide no operator protection and should not be used for applications where operator safety is a concern. Opening and using reagents within a laminar flow hood or dead air box can also help to prevent reagent contamination to reduce waste. The use of PPE, appropriate reagents, engineering controls, along with proper technique can help prevent PCR contamination to ensure results are both accurate and reliable.


